How High Performance Computing is Powering the Age of Genomic Big Data
What does bacteria, a blade of grass and the human body have in common?
On the surface, very little. But given the title of this blog, you’re probably way ahead of me. At a cellular level, they all make use of the same superb genetic database to control their appearance, as well as how they develop, function and behave.
 This incredibly complex information array is encoded into the DNA molecule existing in every cell of every organism. DNA is effectively the manual or blueprint that contains all the essential information and instructions in chemical form to build, grow and maintain a living thing.
This incredibly complex information array is encoded into the DNA molecule existing in every cell of every organism. DNA is effectively the manual or blueprint that contains all the essential information and instructions in chemical form to build, grow and maintain a living thing.
Right now, we are on a voyage of discovery to unlock more of the secrets held within the microscopic structure of DNA. It’s an area of research that has huge potential befits for us all; everything from boosting food production to helping us understand, diagnose, treat and cure a host of diseases.
Why am I particularly interested? Because Linux and High Performance Computing (HPC) are key enabling technologies behind all the research and breakthroughs in this field.
The age of genomic research
All of our genetic information is carried in two strands of DNA (Deoxyribonucleic acid) that coil around each other in a double helix shape. This molecular structure was first correctly identified in 1953 by Francis Crick and James Watson. But it wasn’t until the completion of the Human Genome Project in April 2003 that all the genetic information in the human genome (a technical word for all our DNA) was successfully identified, sequenced and mapped.
This vast international scientific research project took a total of 13 years and cost approximately $2.7 billion to complete. That’s not surprising, because DNA is an amazing and complicated substance.

It has been only 17 years since the initial Human Genome Project was completed. In that time, the cost and time needed to sequence a partial or an entire human genome has been slashed. Today genetic testing has become commonplace and can often be completed in a matter of weeks.
This means we can now easily map the genome of any individual to enable more precise medical treatment. It also makes it possible to build and maintain deep, broad databases of real-world genetic information. With this level of big data, we can begin to automate the large-scale analysis of genetic information helping to improve healthcare for whole populations.
My Key takeaways
There’s no doubt about it. We are living in amazing times. Here are a few parting thoughts I’d like to leave you with:
- The benefits of open source research
The Human Genome Project is an excellent example of large-scale international cooperation. It took a closely-coordinated and collaborative team effort to complete. Once the human genome had been successfully sequenced and decoded, it was immediately made publicly available. Since then, new information has been regularly published and made freely available. Here at SUSE, we’re totally committed to this community-driven “open source” ideal. It permeates everything we do.
- DNA is amazing for data storage
This year, it’s estimated that for every person on the planet, 1.7MB of data will be produced every second. That’s a staggering amount of new data being generated every year. It also means we have a data storage problem looming on the horizon. Fortunately, DNA makes it possible to accurately store vast amounts of data at a density way beyond any electronic devices we’re currently using. It’s also stable and incredibly energy efficient. It might sound like science fiction right now, but just last year all the information on Wikipedia was successfully encoded into synthetic DNA. So, watch this space…
- Economics and performance improvements from HPC solutions
The initial Human Genome Project between 1990 and 2003 was a bioinformatics triumph. This is a multi-discipline field of science that combines biology, computer science, software tools, information engineering, mathematics and statistics. For obvious reasons, the project required the use of supercomputers to handle all the number-crunching involved. As you can see from the graph below, the cost of sequencing human genomes has been drastically dropping over the years.
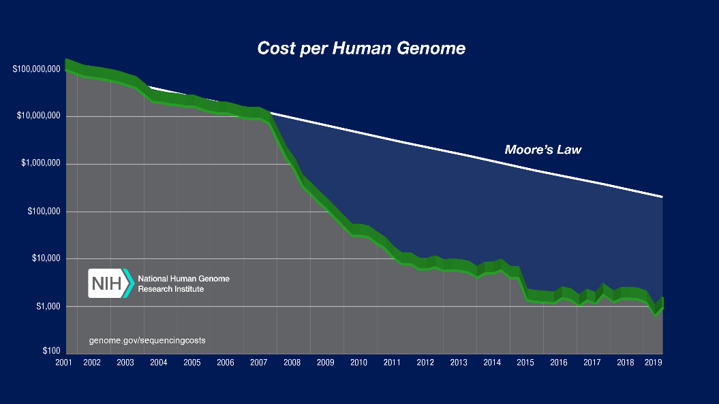
Picture source: National Human Genome Research Institute
There are two reasons for this. One is due to advancements in gene sequencing methods, with more automation and higher throughputs. The other is down to the vastly improved performance and economics of supercomputers. The fastest option today is performing 85 times faster than any option available a decade ago. And every one of the top 500 now runs on Linux, ensuring that it is way more cost-effective.
SUSE leads the way in tailoring Linux for HPC environments. Why not take a moment to check out the information available at either of these links:
- HPC and Advanced Analytics: Three Things You Need to Know
- SUSE Linux Enterprise High Performance Computing
Thanks for reading!
Related Articles
Jul 13th, 2022
Harvesting the Benefits of Cloud-Native Hyperconvergence
Feb 24th, 2023
No comments yet